Next generation sequencing (NGS) is a powerful diagnostic tool for identifying tumor variants. Tumor sequencing workflows often rely on off-the-shelf panels, which allow for multiplexed amplification of known cancer mutational “hotspots”. These panels are diagnostically useful, but they can’t keep pace with the cancer research community’s diligent uncovering of new, previously unknown hotspots, genes, and biomarkers. That’s why scientists at Columbia University Medical Center turned to a customized cancer gene panel from Pillar Biosciences. Using Pillar’s SLIMamp® technology, they identified clinically relevant variants with less than 2.5% allele frequency using as little as 2.5ng of input DNA.
Dr. Helen Fernandes, an associate professor at Columbia University Medical Center, leads the Laboratory of Personalized Genomics Medicine, which validates and develops diagnostic assays for clinical use. Her lab typically runs around 100 clinical NGS assays a week, and the CLIA-accredited lab was in the market for a new assay to detect solid tumor cancer genes.
They were looking for a tool that could detect markers relevant for clinical trials; not only hotspots, but also entire coding sequences along with biomarkers that predict responses to immunotherapy. A robust workflow was also important. Panels used by the lab in the past required reflex testing of at least 15% of cytology specimens to real time PCR because of low DNA quantity, said Dr. Fernandes in a talk at the AMP annual meeting last year.
Dr. Fernandes and her team selected Pillar’s technology for five reasons:
- Panel customization
- Low DNA input
- Single-tube protocol
- Simplified workflow
- Improved turnaround time
Pillar’s Stem-Loop Inhibition Mediated Amplification (SLIMamp) technology uses tagged primers that cause undesirable amplicons to form stem loops that prevent further amplification. The SLIM acronym is fitting, because it lends itself to a simple protocol that relies on less DNA and only three hours of hands-on library prep time (eight hours total). The workflow enables multiplexing of over 1000 variants in a single tube at room temperature. That means fewer man-hours, decreased likelihood of technician errors, and opportunities for automation.
Dr. Fernandes and her team first evaluated the technology using one of Pillar’s off-the-shelf ONCO/Reveal™ panels and 56 previously tested formalin-fixed paraffin-embedded (FFPE) samples harboring 26 clinically relevant variants in 9 genes. Her team tested their ability to detect known variants using the Pillar assay against a TruSeq Amplicon Cancer Panel (Illumina, now discontinued).
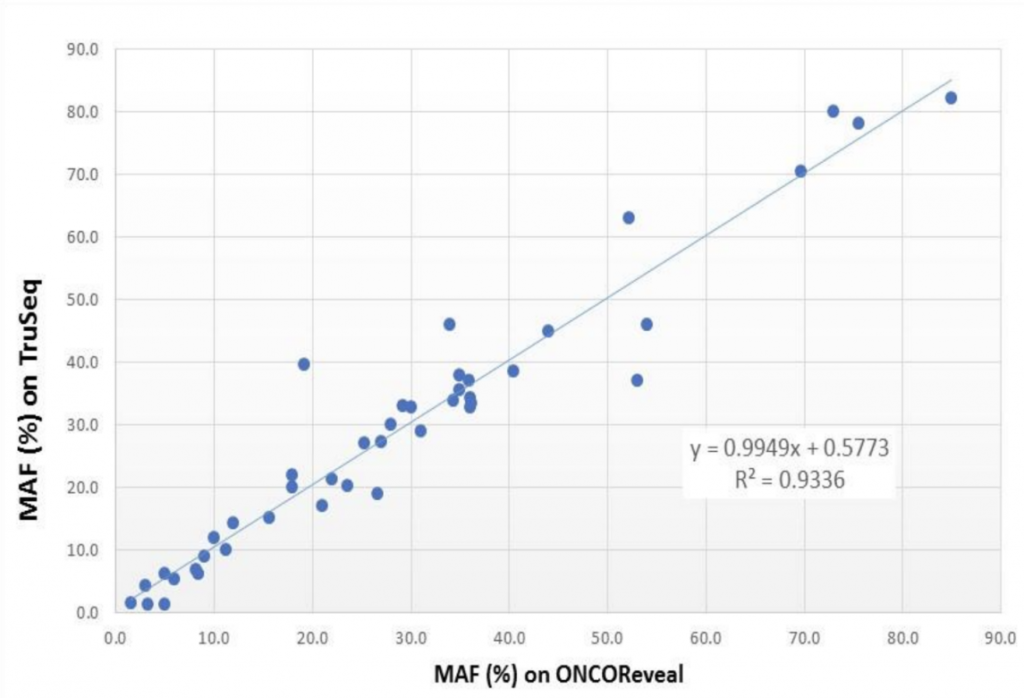
As seen in the graph below, the mutant allele fractions (MAFs) ranged from 3-80% across both the Pillar and Illumina panels with high correlation (R2 = 0.94).
The ONCO/Reveal panel yielded consistent results whether 20, 10, or 2.5ng of DNA was used, even at only 3% MAF. Dr. Fernandes also noted that five samples that were unable to be evaluated with the Illumina TruSeq Panel were successfully sequenced with the Pillar ONCO/Reveal Panel.
After validating the ONCO/Reveal panel against a trusted industry standard, the lab validated a Pillar custom panel consisting of 47 genes with particular therapeutic, prognostic, or diagnostic relevance. The outcomes were comparable to the off-the-shelf panels in terms of MAF percentages and tolerance of low-input DNA.
Dr. Fernandes’ story illustrates the potential of Pillar custom panels as an effective alternative to other NGS panels and a great way to develop an assay to fit your exact design criteria. As clinical researchers at Columbia and elsewhere identify new markers, the flexibility of Pillar custom panels and simplicity of the SLIMamp workflow will continue to accelerate cancer research into the future.

