The cancer research community relies on formalin-fixed samples, but the quality of these samples degrades over time. Sequencing data from partially degraded samples is plagued with artifacts. To address this “garbage in, garbage out” problem, Pillar Biosciences has developed a suite of tools that help sort actual variants from artifactual variants in low quality samples.
Formalin-Fixed Paraffin Embedding (FFPE) is a sample preservation method used in genetics and cell biology research to store tissue samples at room temperature over long periods of time. While FFPE is a cost-effective method for long-term sample storage, formalin is a toxic compound known to degrade nucleic acids. That spells trouble for genomic analysis.
The problem with FFPE is false positives. False positives plague variant interpretation in FFPE samples, because degradation by formalin mimics mutations. False positives are particularly prevalent when the input DNA is very minimal, so there are fewer template molecules to amplify.
You can think of this sample noise as “garbage in”. And if you feed garbage data into typical bioinformatics workflows, you’re going to get garbage out; that is, nonsense or misleading results.
Pillar’s technology helps users recover good data from formalin-damaged samples in two ways.
- First, our SLIMamp single-tube library-prep workflow allows for UDG pre-treatment of multiplexed samples, which can repair some of the formalin artifacts.
- Second, our PiVAT bioinformatics software filters errors using a proprietary noise suppression capability.
We evaluated the performance of our ONCO/Reveal Lung and Colon Cancer Panel using Horizon Discovery’s Formalin Compromised Standard Series, which includes mild, moderate, and severely damaged formalin degraded DNA (fcDNA). We used our panels to analyze five allelic mutations of varying frequencies in Horizon’s fcDNA (Figure 1).
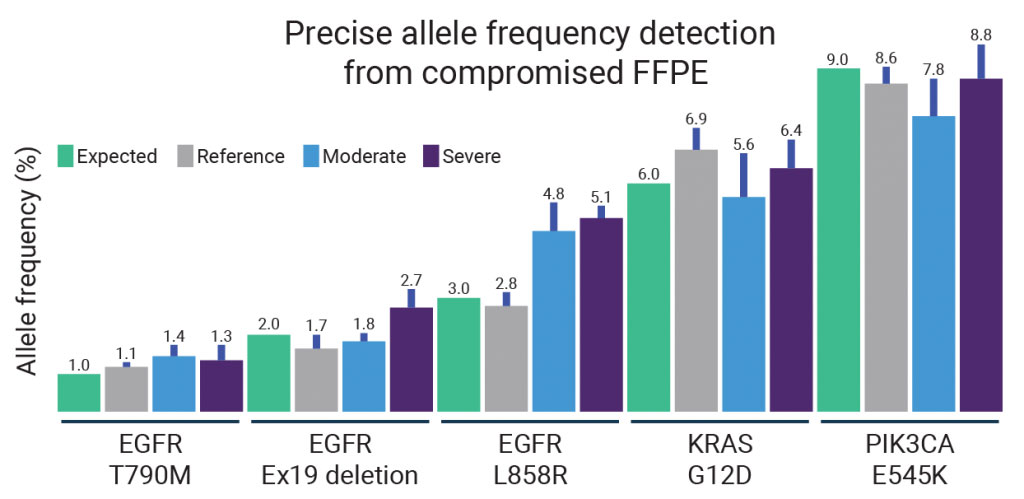
For most mutations, the expected allele frequency fell within the standard deviation of the measured allele frequency, even for severely degraded samples. That means, that for many samples, UDG treatment in our SLIMamp workflow reduces “garbage in”.
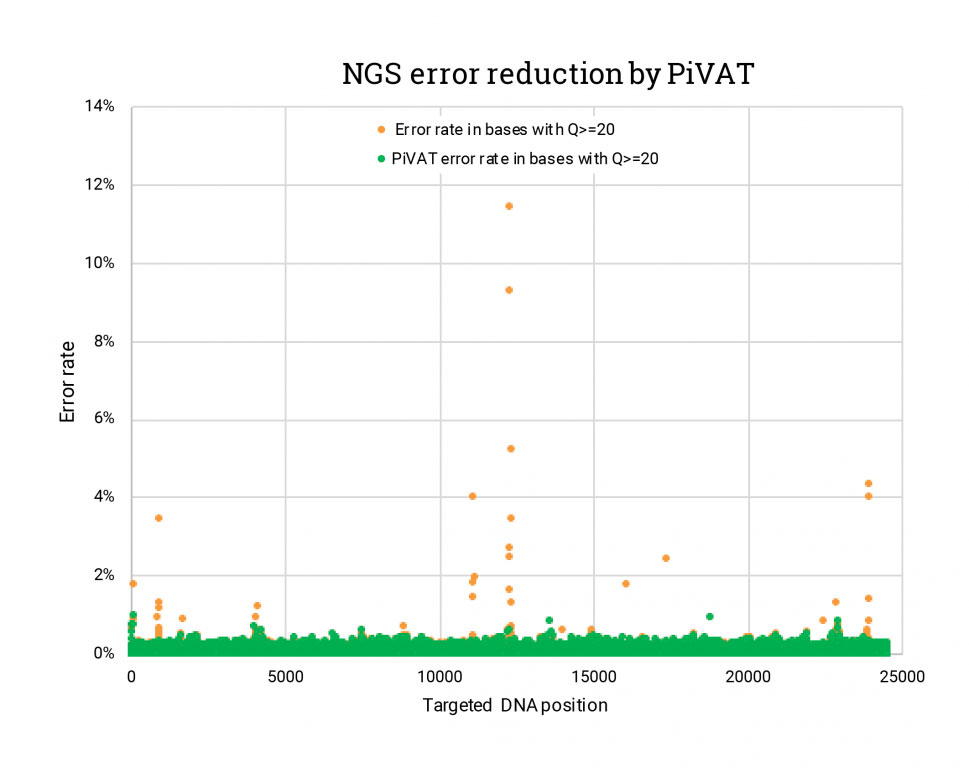
“Garbage out” is a bioinformatics problem, and for that, we developed our Pillar Variant Analysis Toolkit (PiVAT). Variant calls you can trust start with proper assemblies. After an initial assembly, PiVAT performs local realignment and reassembly. Local realignment and proprietary heuristic methods help the PiVAT platform sort out additional noise for cleaner results (Figure 2).
FFPE tissue preservation is an important and cost-effective tool for the cancer research community, and it’s not going anywhere. At Pillar, we’ve developed library preparation and bioinformatic solutions, so that you can make sequencing work for your favorite sample preservation method. To see our technology in action with real clinical FFPE samples, keep reading.

